Biomolecules such as proteins and RNA form large complexes that work together to accomplish core biological functions. Dysfunction of those biomolecules may result in severe diseases. To understand such diseases and develop treatments, mechanisms of these protein functions need to be understood. This requires determining their 3-dimensional structures. However, most biomolecules move and change their shapes upon accomplishing their functions, and such dynamic factors should also be considered.
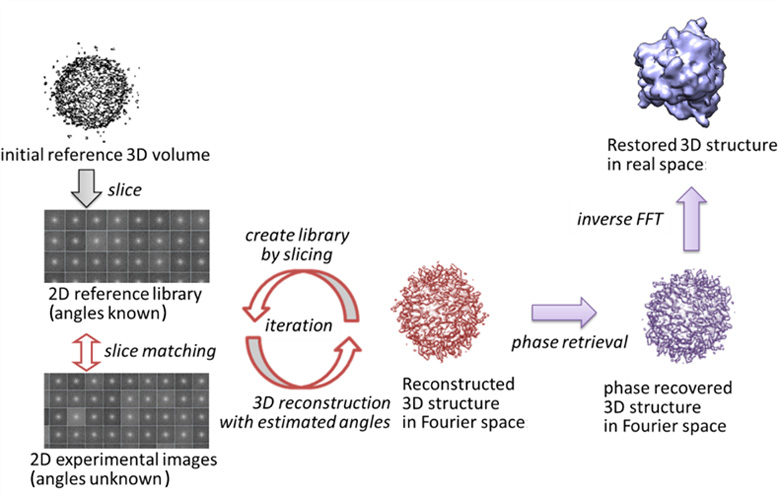
3D structures of biomolecules are obtained primarily through experimental studies. For example, X-ray crystallography provides high-resolution structures. However, it requires crystallization of the biological molecule, which can be quite a challenge. In other methods, such as cryo-electron microscopy (cryo-EM), 2D images of the individual biological molecules are collected, and then be reassembled to form a 3D structure, though generally of a lower resolution. Still, this presents some advantages over X-ray crystallography as large complexes can be studied as well as their dynamics.
Recently, the X-ray free electron laser (XFEL) also opened the possibility to directly image single molecules, using its high intensity X-ray laser. Our interdisciplinary research unit aims to develop computational tools to determine 3D structures and dynamics, utilizing cryo-EM and XFEL experimental data. Such computational tools will be employed on the K and post-K computers to take advantage of the resources for large scale data analyses, and also to share these tools with the scientific community. Applications could provide new insights into the structure and dynamics of important biomolecules that are unattainable with existing techniques.